-
Judith Miller
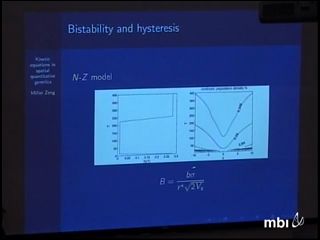
We derive kinetic differential or integrodifference equations for the mean and variance or of a quantitative trait as a function of space and time, in some cases recovering known equations and in some cases obtaining new ones that capture effects, such as nonmonotonicity of traveling waves, that can be seen in stochastic simulations. We then reanalyze kinetic equations due to Kirkpatrick and Barton for population range limits, showing that they exhibit bistability and hysteresis. This suggests a possible mechanism for lag times between establishment and subsequent explosive growth and range expansion in the absence of an Allee effect.
-
Peter Jagers
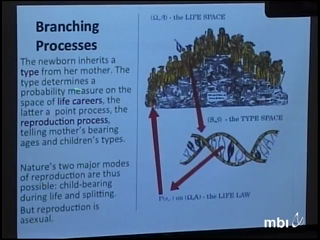
In a toy model of binary splitting branching processes with population size dependence (supercritical below and subcritical above a threshhold, the carrying capacity) the chance of a little population establishing itself in the sense of reaching a band around the carrying capacity is determined, and so is the persistence time of the population. Mutations and competition between morphs are introduced, and it turns out that the resulting processes exhibit evolutionary branchings which occur in a manner slightly different from that predicted by established deterministic theory. The validity of conclusions is discussed in terms of more general branching processes. (Joint work with Serik Sagitov, Fima Klebaner et al.)
-
David Anderson
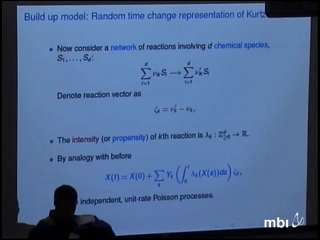
I will focus on computational methods for stochastically modeled biochemical reaction networks. The simplest stochastic models of such networks treat the system as a continuous time Markov chain with the state being the number of molecules of each species and with reactions modeled as possible transitions of the chain. I will show how different computational methods can be understood and analyzed by using different representations for the processes. Topics discussed will be a subset of: approximation techniques, variance reduction methods, parameter sensitivities.
-

Ilya Nemenman
Stochastic biochemical systems and population genetics models are described by similar mathematical equations, and hence similar phenomena should be observed in both systems. Here we focus on stochastic kinetics with time scale separation. We show how to integrate out the fast degrees of freedom, while rigorously preserving their effects on the fluctuations of slower variables. This procedure allows to speed up simulation of kinetic networks and reveals a number of interesting phenomena, previously unobserved in the context of classical stochastic kinetics. One of the most interesting is the emergence of geometric phases, which we show may have substantial effects on, in particular, the frequency of fixation of new mutations in slowly variable environments.
-

Johan Metz
Deterministic population dynamical models connect to reality through their interpretation as limits for systems size going to infinity of stochastic processes in which individuals are represented as discrete entities. In structured population models individuals may be born in different states (e.g. locations in space) after which they proceed through their h(eterogeneity)-state space, e.g. spanned by their i(dividual)-state and location. On such models one can graft evolutionary processes like random genetic drift or adaptive evolution by rare repeated substitutions of mutants in heritable traits affecting the state transition and reproduction processes of individuals. From this general perspective I will derive the so-called Canonical Equation of adaptive dynamics, a differential equation for evolutionary trait change derived under the additional assumption that mutations have small effect. In the CE approximation the rate of evolution is found to correspond to the product of a parameter $n_{e,A}$, equal to the population size times a dimensionless product of life history parameters (including spatial movements), times the gradient of the invasion fitness of potential mutants with respect to their trait vector. From a heuristic connection with the diffusion approximation for genetic drift it follows that $n_{e,A} = n_{e,D}$, the effective population size from population genetics.
-

Linda Allen
In deterministic epidemic models, pathogen extinction in a population is determined by the magnitude of the basic reproduction number R0. In stochastic epidemic models, the probability of pathogen extinction depends on R0, the size of the population and the number of infectious individuals. For example, in the SIS Markov chain epidemic model, if the basic reproduction number R0>1, the population size is large and I(0)=a is small, then a classic result of Whittle (1955) gives an approximation to the probability of pathogen extinction: (1/R0)a. This classic result can be derived from branching process theory. We apply results from multitype Markov branching process theory to generalize this approximation for probability of pathogen extinction to more complex epidemic models with multiple stages, treatment , or multiple populations and to within host models of virus and cell dynamics.
Work done in collaboration with Yuan Yuan and Glenn Lahodny.
-
Peter Kramer
Recent years have seen increasing attention to the subtle effects on intracellular transport caused when multiple molecular motors bind to a common cargo. We develop and examine a coarse-grained model which resolves the spatial configuration as well as the thermal fluctuations of the molecular motors and the cargo. This intermediate model can accept as inputs either common experimental quantities or the effective single-motor transport characterizations obtained through the kind of systematic analysis of detailed molecular motor models described in Fricks' presentation. Through stochastic asymptotic reductions, we derive the effective transport properties of the multiple-motor-cargo complex, and provide analytical explanations for why a cargo bound to two molecular motors moves more slowly at low applied forces but more rapidly at high applied forces than a cargo bound to a single molecular motor.
-
John Fricks
A stochastic model for variable-length stepping of kinesins engineered with extended neck linkers is developed. This requires consideration of the separation in microtubule binding sites between the heads of the motor at the beginning of a step. It can be shown that the separation is a stationary Markov process and can be included in the calculation of standard experimental quantities, such as asymptotic velocity and effective diffusion, through the appropriate limits of a semi-Markov process. Using this framework, asymptotic results for randomly detached motors are also obtained and linked to the statistical analysis of velocity data from motor assays. In addition, we will discuss how the framework developed here could be used as one component of a larger scale model for motor-cargo systems of the type to be presented in Kramer's talk.
-
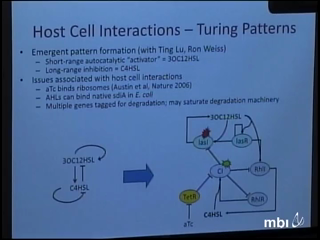
David Karig
The growing field of synthetic biology aims to forward engineer biology both for applications such as energy production, drug production, and bioremediation, as well as for the purpose of furthering the fundamental understanding of natural systems. However, engineering living cells is notoriously difficult due to issues such as mutation, epigenetic variation, fitness effects, and the interaction of synthetic components with host cell processes. Thus, simpler contexts such as cell-free expression systems offer great promise to engineering complex biological behavior in a quantitative fashion. Furthermore, the confinement of cell-free gene circuit reactions in nanofabricated reaction devices offers a flexible approach to investigating fundamental aspects of gene circuit function. Currently, we are using such devices to study noise in simple gene circuits. Cell-free reactions confined in different volume wells are imaged over time using fluorescent microscopy. The noise characteristics of the resulting gene expression trajectories are analyzed and compared for different gene circuits.
-

Ellen Baake
I will start with an overview over various models for the dynamics of the genetic composition of populations evolving under recombination. For the deterministic treatment that applies in the infinite-population limit, one has large, nonlinear dynamical systems; for the stochastic treatment required for finite populations, the Moran, or Wright-Fisher model is appropriate.
I will focus on models involving only single crossovers in every generation and contrast the situations in continuous and in discrete time. In continuous time, the deterministic model has a simple closed solution, which is due to the independence of the individual recombination events. In contrast, discrete time introduces dependencies between the links and leads to a much more complex situation. Nevertheless, the situation becomes tractable by looking backwards in time, starting from single individuals at present in a Wright-Fisher population with recombination and tracing back the ancestry of the various gene segments that result from recombination. In the limit of population size to infinity, these segments become independent. We identify the process that describes their history, together with the tree structures they define, which we like to call ancestral recombination trees. It turns out that the corresponding tree _topologies_ play a special role: Surprisingly, explicit probabilities may be assigned to them, which then leads to an explicit solution of the recombination dynamics.
This is joint work with Ute von Wangenheim.
[1] E. Baake, Deterministic and stochastic aspects of single-crossover recombination, Proceedings of the International Congress of Mathematicians, Hyderabad, India, 2010, Vol. VI, 3037-3053
-
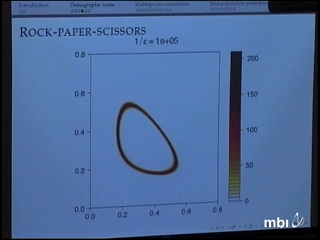
Sebastian Schreiber
Populations, whether they be viral particles, bio-chemicals, plants or animals, are subject to intrinsic and extrinsic sources of stochasticity. This stochasticity in conjunction with nonlinear interactions between individuals determines to what extinct populations are able to persist in the long-term. Understanding the precise nature of these interactive effects is a central issue in population biology from theoretical, empirical, and applied perspectives.
For the first part of this talk, I will discuss, briefly, the relationship between attractors of deterministic models and quasi-stationary distributions of their stochastic, finite population counterpoints i.e. models accounting for demographic stochasticity. These results shed some insight into when persistence should be observed over long time frames despite extinction being inevitable.
For the second part of the talk, I will discuss results on stochastic persistence and boundedness for stochastic models accounting for environmental (but not demographic) noise. Stochastic boundedness asserts that asymptotically the population process tends to remain in compact sets. In contrast, stochastic persistence requires that the population process tends to be "repelled" by some "extinction set." Using these results, I will illustrate how environmental noise can facilitate coexistence of competing species and how dispersal in stochastic environments can rescue locally extinction prone populations. Empirical demonstrations from Kansas prairies, acorn woodpecker populations, and microcosm experiments will be discussed.
-
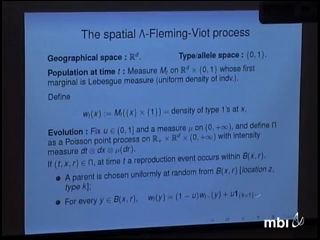
Amandine Veber
The SLFV process is a population model in which individuals live in a continuous space. Each of them also carries some heritable type or allele. We shall describe the long-term behaviour of this measure-valued process and that of the corresponding genealogical process of a sample of individuals in two cases : one that mimics the evolution of nearest-neighbour voter model (but in a spatial continuum), and one that allows some individuals to send offspring at very large distances. This is a joint work with Nathana�l Berestycki and Alison Etheridge.
-
John Yin
The dynamics of a virus infection within its host is governed at its earliest stages by processes at the molecular and cellular scale. We are developing cell-culture measurements and computational models to better understand how these and other processes contribute to the early dynamics of virus growth and infection spread. As a model system we study vesicular stomatitis virus (VSV), a rabies-like RNA virus, growing on BHK cells. Established single-cycle measures of virus growth within infected cells provide population averages, which mask potential cell-to-cell variation. We used fluorescence-activated cell sorting to isolate single cells infected by single particles of a recombinant VSV expressing green fluorescent protein. Measured virus yields spanned a broad range from 8000 to below the detection limit of 10 infectious virus particles per cell. Viral genetic variation and host-cell cycle differences were unable to fully account for the observed yield differences. Computer simulations of the VSV dynamics within an infected cell suggest a potential role for stochastic gene expression to the observed yield variation. These studies are currently being extended to study the kinetics of virus production from individual infected cells.
 Judith MillerWe derive kinetic differential or integrodifference equations for the mean and variance or of a quantitative trait as a function of space and time, in some cases recovering known equations and in some cases obtaining new ones that capture effects, such as nonmonotonicity of traveling waves, that can be seen in stochastic simulations. We then reanalyze kinetic equations due to Kirkpatrick and Barton for population range limits, showing that they exhibit bistability and hysteresis. This suggests a possible mechanism for lag times between establishment and subsequent explosive growth and range expansion in the absence of an Allee effect.
Judith MillerWe derive kinetic differential or integrodifference equations for the mean and variance or of a quantitative trait as a function of space and time, in some cases recovering known equations and in some cases obtaining new ones that capture effects, such as nonmonotonicity of traveling waves, that can be seen in stochastic simulations. We then reanalyze kinetic equations due to Kirkpatrick and Barton for population range limits, showing that they exhibit bistability and hysteresis. This suggests a possible mechanism for lag times between establishment and subsequent explosive growth and range expansion in the absence of an Allee effect. Peter JagersIn a toy model of binary splitting branching processes with population size dependence (supercritical below and subcritical above a threshhold, the carrying capacity) the chance of a little population establishing itself in the sense of reaching a band around the carrying capacity is determined, and so is the persistence time of the population. Mutations and competition between morphs are introduced, and it turns out that the resulting processes exhibit evolutionary branchings which occur in a manner slightly different from that predicted by established deterministic theory. The validity of conclusions is discussed in terms of more general branching processes. (Joint work with Serik Sagitov, Fima Klebaner et al.)
Peter JagersIn a toy model of binary splitting branching processes with population size dependence (supercritical below and subcritical above a threshhold, the carrying capacity) the chance of a little population establishing itself in the sense of reaching a band around the carrying capacity is determined, and so is the persistence time of the population. Mutations and competition between morphs are introduced, and it turns out that the resulting processes exhibit evolutionary branchings which occur in a manner slightly different from that predicted by established deterministic theory. The validity of conclusions is discussed in terms of more general branching processes. (Joint work with Serik Sagitov, Fima Klebaner et al.) David AndersonI will focus on computational methods for stochastically modeled biochemical reaction networks. The simplest stochastic models of such networks treat the system as a continuous time Markov chain with the state being the number of molecules of each species and with reactions modeled as possible transitions of the chain. I will show how different computational methods can be understood and analyzed by using different representations for the processes. Topics discussed will be a subset of: approximation techniques, variance reduction methods, parameter sensitivities.
David AndersonI will focus on computational methods for stochastically modeled biochemical reaction networks. The simplest stochastic models of such networks treat the system as a continuous time Markov chain with the state being the number of molecules of each species and with reactions modeled as possible transitions of the chain. I will show how different computational methods can be understood and analyzed by using different representations for the processes. Topics discussed will be a subset of: approximation techniques, variance reduction methods, parameter sensitivities. Ilya NemenmanStochastic biochemical systems and population genetics models are described by similar mathematical equations, and hence similar phenomena should be observed in both systems. Here we focus on stochastic kinetics with time scale separation. We show how to integrate out the fast degrees of freedom, while rigorously preserving their effects on the fluctuations of slower variables. This procedure allows to speed up simulation of kinetic networks and reveals a number of interesting phenomena, previously unobserved in the context of classical stochastic kinetics. One of the most interesting is the emergence of geometric phases, which we show may have substantial effects on, in particular, the frequency of fixation of new mutations in slowly variable environments.
Ilya NemenmanStochastic biochemical systems and population genetics models are described by similar mathematical equations, and hence similar phenomena should be observed in both systems. Here we focus on stochastic kinetics with time scale separation. We show how to integrate out the fast degrees of freedom, while rigorously preserving their effects on the fluctuations of slower variables. This procedure allows to speed up simulation of kinetic networks and reveals a number of interesting phenomena, previously unobserved in the context of classical stochastic kinetics. One of the most interesting is the emergence of geometric phases, which we show may have substantial effects on, in particular, the frequency of fixation of new mutations in slowly variable environments. Johan MetzDeterministic population dynamical models connect to reality through their interpretation as limits for systems size going to infinity of stochastic processes in which individuals are represented as discrete entities. In structured population models individuals may be born in different states (e.g. locations in space) after which they proceed through their h(eterogeneity)-state space, e.g. spanned by their i(dividual)-state and location. On such models one can graft evolutionary processes like random genetic drift or adaptive evolution by rare repeated substitutions of mutants in heritable traits affecting the state transition and reproduction processes of individuals. From this general perspective I will derive the so-called Canonical Equation of adaptive dynamics, a differential equation for evolutionary trait change derived under the additional assumption that mutations have small effect. In the CE approximation the rate of evolution is found to correspond to the product of a parameter $n_{e,A}$, equal to the population size times a dimensionless product of life history parameters (including spatial movements), times the gradient of the invasion fitness of potential mutants with respect to their trait vector. From a heuristic connection with the diffusion approximation for genetic drift it follows that $n_{e,A} = n_{e,D}$, the effective population size from population genetics.
Johan MetzDeterministic population dynamical models connect to reality through their interpretation as limits for systems size going to infinity of stochastic processes in which individuals are represented as discrete entities. In structured population models individuals may be born in different states (e.g. locations in space) after which they proceed through their h(eterogeneity)-state space, e.g. spanned by their i(dividual)-state and location. On such models one can graft evolutionary processes like random genetic drift or adaptive evolution by rare repeated substitutions of mutants in heritable traits affecting the state transition and reproduction processes of individuals. From this general perspective I will derive the so-called Canonical Equation of adaptive dynamics, a differential equation for evolutionary trait change derived under the additional assumption that mutations have small effect. In the CE approximation the rate of evolution is found to correspond to the product of a parameter $n_{e,A}$, equal to the population size times a dimensionless product of life history parameters (including spatial movements), times the gradient of the invasion fitness of potential mutants with respect to their trait vector. From a heuristic connection with the diffusion approximation for genetic drift it follows that $n_{e,A} = n_{e,D}$, the effective population size from population genetics. Linda AllenIn deterministic epidemic models, pathogen extinction in a population is determined by the magnitude of the basic reproduction number R0. In stochastic epidemic models, the probability of pathogen extinction depends on R0, the size of the population and the number of infectious individuals. For example, in the SIS Markov chain epidemic model, if the basic reproduction number R0>1, the population size is large and I(0)=a is small, then a classic result of Whittle (1955) gives an approximation to the probability of pathogen extinction: (1/R0)a. This classic result can be derived from branching process theory. We apply results from multitype Markov branching process theory to generalize this approximation for probability of pathogen extinction to more complex epidemic models with multiple stages, treatment , or multiple populations and to within host models of virus and cell dynamics.
Linda AllenIn deterministic epidemic models, pathogen extinction in a population is determined by the magnitude of the basic reproduction number R0. In stochastic epidemic models, the probability of pathogen extinction depends on R0, the size of the population and the number of infectious individuals. For example, in the SIS Markov chain epidemic model, if the basic reproduction number R0>1, the population size is large and I(0)=a is small, then a classic result of Whittle (1955) gives an approximation to the probability of pathogen extinction: (1/R0)a. This classic result can be derived from branching process theory. We apply results from multitype Markov branching process theory to generalize this approximation for probability of pathogen extinction to more complex epidemic models with multiple stages, treatment , or multiple populations and to within host models of virus and cell dynamics. Peter KramerRecent years have seen increasing attention to the subtle effects on intracellular transport caused when multiple molecular motors bind to a common cargo. We develop and examine a coarse-grained model which resolves the spatial configuration as well as the thermal fluctuations of the molecular motors and the cargo. This intermediate model can accept as inputs either common experimental quantities or the effective single-motor transport characterizations obtained through the kind of systematic analysis of detailed molecular motor models described in Fricks' presentation. Through stochastic asymptotic reductions, we derive the effective transport properties of the multiple-motor-cargo complex, and provide analytical explanations for why a cargo bound to two molecular motors moves more slowly at low applied forces but more rapidly at high applied forces than a cargo bound to a single molecular motor.
Peter KramerRecent years have seen increasing attention to the subtle effects on intracellular transport caused when multiple molecular motors bind to a common cargo. We develop and examine a coarse-grained model which resolves the spatial configuration as well as the thermal fluctuations of the molecular motors and the cargo. This intermediate model can accept as inputs either common experimental quantities or the effective single-motor transport characterizations obtained through the kind of systematic analysis of detailed molecular motor models described in Fricks' presentation. Through stochastic asymptotic reductions, we derive the effective transport properties of the multiple-motor-cargo complex, and provide analytical explanations for why a cargo bound to two molecular motors moves more slowly at low applied forces but more rapidly at high applied forces than a cargo bound to a single molecular motor. John FricksA stochastic model for variable-length stepping of kinesins engineered with extended neck linkers is developed. This requires consideration of the separation in microtubule binding sites between the heads of the motor at the beginning of a step. It can be shown that the separation is a stationary Markov process and can be included in the calculation of standard experimental quantities, such as asymptotic velocity and effective diffusion, through the appropriate limits of a semi-Markov process. Using this framework, asymptotic results for randomly detached motors are also obtained and linked to the statistical analysis of velocity data from motor assays. In addition, we will discuss how the framework developed here could be used as one component of a larger scale model for motor-cargo systems of the type to be presented in Kramer's talk.
John FricksA stochastic model for variable-length stepping of kinesins engineered with extended neck linkers is developed. This requires consideration of the separation in microtubule binding sites between the heads of the motor at the beginning of a step. It can be shown that the separation is a stationary Markov process and can be included in the calculation of standard experimental quantities, such as asymptotic velocity and effective diffusion, through the appropriate limits of a semi-Markov process. Using this framework, asymptotic results for randomly detached motors are also obtained and linked to the statistical analysis of velocity data from motor assays. In addition, we will discuss how the framework developed here could be used as one component of a larger scale model for motor-cargo systems of the type to be presented in Kramer's talk. David KarigThe growing field of synthetic biology aims to forward engineer biology both for applications such as energy production, drug production, and bioremediation, as well as for the purpose of furthering the fundamental understanding of natural systems. However, engineering living cells is notoriously difficult due to issues such as mutation, epigenetic variation, fitness effects, and the interaction of synthetic components with host cell processes. Thus, simpler contexts such as cell-free expression systems offer great promise to engineering complex biological behavior in a quantitative fashion. Furthermore, the confinement of cell-free gene circuit reactions in nanofabricated reaction devices offers a flexible approach to investigating fundamental aspects of gene circuit function. Currently, we are using such devices to study noise in simple gene circuits. Cell-free reactions confined in different volume wells are imaged over time using fluorescent microscopy. The noise characteristics of the resulting gene expression trajectories are analyzed and compared for different gene circuits.
David KarigThe growing field of synthetic biology aims to forward engineer biology both for applications such as energy production, drug production, and bioremediation, as well as for the purpose of furthering the fundamental understanding of natural systems. However, engineering living cells is notoriously difficult due to issues such as mutation, epigenetic variation, fitness effects, and the interaction of synthetic components with host cell processes. Thus, simpler contexts such as cell-free expression systems offer great promise to engineering complex biological behavior in a quantitative fashion. Furthermore, the confinement of cell-free gene circuit reactions in nanofabricated reaction devices offers a flexible approach to investigating fundamental aspects of gene circuit function. Currently, we are using such devices to study noise in simple gene circuits. Cell-free reactions confined in different volume wells are imaged over time using fluorescent microscopy. The noise characteristics of the resulting gene expression trajectories are analyzed and compared for different gene circuits. Ellen BaakeI will start with an overview over various models for the dynamics of the genetic composition of populations evolving under recombination. For the deterministic treatment that applies in the infinite-population limit, one has large, nonlinear dynamical systems; for the stochastic treatment required for finite populations, the Moran, or Wright-Fisher model is appropriate.
Ellen BaakeI will start with an overview over various models for the dynamics of the genetic composition of populations evolving under recombination. For the deterministic treatment that applies in the infinite-population limit, one has large, nonlinear dynamical systems; for the stochastic treatment required for finite populations, the Moran, or Wright-Fisher model is appropriate. Sebastian SchreiberPopulations, whether they be viral particles, bio-chemicals, plants or animals, are subject to intrinsic and extrinsic sources of stochasticity. This stochasticity in conjunction with nonlinear interactions between individuals determines to what extinct populations are able to persist in the long-term. Understanding the precise nature of these interactive effects is a central issue in population biology from theoretical, empirical, and applied perspectives.
Sebastian SchreiberPopulations, whether they be viral particles, bio-chemicals, plants or animals, are subject to intrinsic and extrinsic sources of stochasticity. This stochasticity in conjunction with nonlinear interactions between individuals determines to what extinct populations are able to persist in the long-term. Understanding the precise nature of these interactive effects is a central issue in population biology from theoretical, empirical, and applied perspectives. Amandine VeberThe SLFV process is a population model in which individuals live in a continuous space. Each of them also carries some heritable type or allele. We shall describe the long-term behaviour of this measure-valued process and that of the corresponding genealogical process of a sample of individuals in two cases : one that mimics the evolution of nearest-neighbour voter model (but in a spatial continuum), and one that allows some individuals to send offspring at very large distances. This is a joint work with Nathana�l Berestycki and Alison Etheridge.
Amandine VeberThe SLFV process is a population model in which individuals live in a continuous space. Each of them also carries some heritable type or allele. We shall describe the long-term behaviour of this measure-valued process and that of the corresponding genealogical process of a sample of individuals in two cases : one that mimics the evolution of nearest-neighbour voter model (but in a spatial continuum), and one that allows some individuals to send offspring at very large distances. This is a joint work with Nathana�l Berestycki and Alison Etheridge. John YinThe dynamics of a virus infection within its host is governed at its earliest stages by processes at the molecular and cellular scale. We are developing cell-culture measurements and computational models to better understand how these and other processes contribute to the early dynamics of virus growth and infection spread. As a model system we study vesicular stomatitis virus (VSV), a rabies-like RNA virus, growing on BHK cells. Established single-cycle measures of virus growth within infected cells provide population averages, which mask potential cell-to-cell variation. We used fluorescence-activated cell sorting to isolate single cells infected by single particles of a recombinant VSV expressing green fluorescent protein. Measured virus yields spanned a broad range from 8000 to below the detection limit of 10 infectious virus particles per cell. Viral genetic variation and host-cell cycle differences were unable to fully account for the observed yield differences. Computer simulations of the VSV dynamics within an infected cell suggest a potential role for stochastic gene expression to the observed yield variation. These studies are currently being extended to study the kinetics of virus production from individual infected cells.
John YinThe dynamics of a virus infection within its host is governed at its earliest stages by processes at the molecular and cellular scale. We are developing cell-culture measurements and computational models to better understand how these and other processes contribute to the early dynamics of virus growth and infection spread. As a model system we study vesicular stomatitis virus (VSV), a rabies-like RNA virus, growing on BHK cells. Established single-cycle measures of virus growth within infected cells provide population averages, which mask potential cell-to-cell variation. We used fluorescence-activated cell sorting to isolate single cells infected by single particles of a recombinant VSV expressing green fluorescent protein. Measured virus yields spanned a broad range from 8000 to below the detection limit of 10 infectious virus particles per cell. Viral genetic variation and host-cell cycle differences were unable to fully account for the observed yield differences. Computer simulations of the VSV dynamics within an infected cell suggest a potential role for stochastic gene expression to the observed yield variation. These studies are currently being extended to study the kinetics of virus production from individual infected cells.